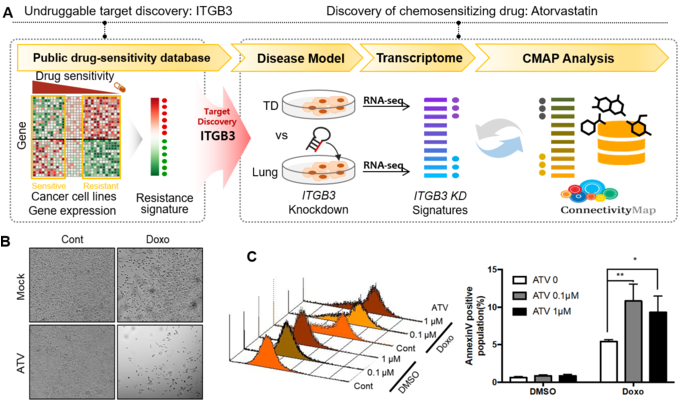

Even when targets responsible for chemoresistance are identified, drug development is often hampered due to the poor druggability of these proteins. We systematically analyzed therapy-resistance with a large-scale cancer cell transcriptome and drug-response datasets and predicted the candidate drugs based on the gene expression profile. Our results implicated the epithelial–mesenchymal transition as a common mechanism underlying resistance to chemotherapeutic drugs. Notably, we identified ITGB3, whose expression was abundant in both drug resistance and mesenchymal status, as a promising target to overcome chemoresistance. We also confirmed that depletion of ITGB3 sensitized cancer cells to conventional chemotherapeutic drugs by modulating the NF-κB signaling pathway. Considering the poor druggability of ITGB3 and the lack of feasible drugs to directly inhibit this protein, we took an in silico screening for drugs mimicking the transcriptome-level changes caused by knockdown of ITGB3. This approach successfully identified atorvastatin as a novel candidate for drug repurposing, paving an alternative path to drug screening that is applicable to undruggable targets.
Soon-Ki Hong*, Haeseung Lee*, Ok-Seon Kwon*, Na-Young Song, Hyo-Ju Lee, Seungmin Kan, Jeong-Hwan Kim, Mirang Kim, Wankyu Kim, Hyuk-Jin Cha, Large-scale pharmacogenomics based drug discovery for ITGB3 dependent chemoresistance in mesenchymal lung cancer, Mol. Cancer 17:175. doi: 10.1186/s12943-018-0924-8. (2018) IF:11.350